The Multiscale Molecular Microscope
TR&D Project 1. The Sample Stage: Tools for Isolating and Preserving Macromolecular Hierarchies
Any given macromolecule may make stable, dynamic or transient interactions with other macromolecules. These interactions form a hierarchical network, comprising a complex series of interactions called the cellular interactome. This whole network is surrounded by a macromolecular milieu of other complexes that jostle with it, and can form vicinal interactions. TR&D1 represents the first module of our pipeline, where the challenge is to preserve with high fidelity and isolate the local native intracellular macromolecular environment of any macromolecule of interest.
To access and then isolate a macromolecule and its interactors in their native hierarchical assemblies, we must break open the cell. However, this disrupts the cell’s large-scale organization; and factors such as dilution, change in milieu, the intermingling of normally segregated components, loss of energy regeneration, and exposure to degradative enzymes cause disassembly, scrambling and aggregation. All these factors worsen as the time lengthens between cell disruption and analytic isolation of the assemblies. Our solution is both to capture as rapidly and efficiently as possible these assemblies, and preserve their native state as much as possible.
To accomplish this, TR&D1 is split into two goals:
Goal 1: Produce Optimized Reagents for Ultra-Efficient Detection and Capture of Complexes from cells.
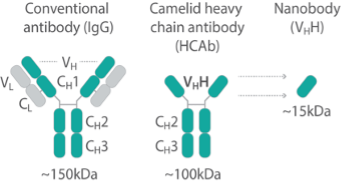
Develop baits for both widely-used existing tags and targets for which the addition of tags are not practical
Generating broadly useful nanobodies has been a priority for the NCDIR, so we first generated nanobodies against common tags. In response to numerous community requests, we have made nanobody repertoires against 12 different targets that for diverse reasons are difficult to tag (protozoan parasites, viruses, transposons, chromatin proteins etc.).
Goal 2: Generate tools and methodologies to preserve these complexes.
Ultrafast Preservation and Preparation Methods for Endogenous Complexes.
A central concept of our Center is rapid preservation and access to any macromolecular complex in its native state by literally freezing the complexes inside the cell, placing them in a state of “suspended animation”, or by adding cell-permeable linkers to instantly chemically preserve the complex upon addition. We have adapted EM technologies for freezing cells and tissues to minimize cellular damage and we explore complementary chemical stabilization methods that compatible with our approaches. To access macromolecular complexes in frozen cells while preserving them, we have developed our cryomilling methods. Our goal is to produce particles with diameters so small, on the order of a few tens of nm, that they are only a little larger than the complexes themselves. To learn more about the cryo-milling technique, take a look at our protocols page!
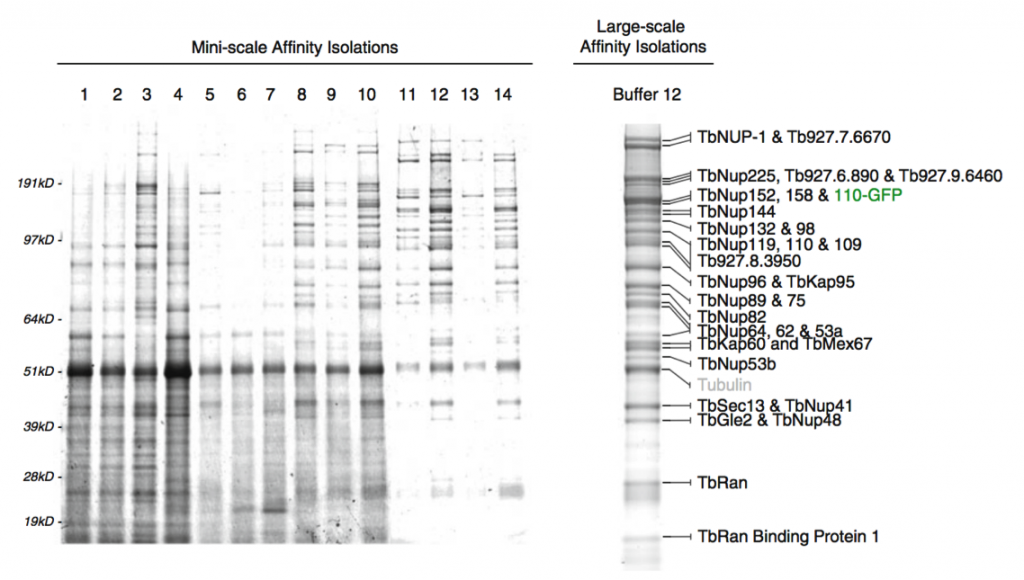
Affinity capture of the Trypanosome nuclear pore complex (Obado et al, 2016)
The methods we develop will enable means for rapidly optimizing these affinity captures, which are both portable to (i) bench scale for any other researcher and (ii) readily accessible, affordable robotic platforms to maximize throughput and reproducibility. Because these technologies must be readily accessible to any laboratory, we are focusing on establishing simple, robust and widely used reagents and approaches for the efficient and rapid isolation of macromolecular complexes. We wish to extend this progress and develop enabling new technologies to allow any researcher to readily and comprehensively access the information in the dynamic interactomes of both normal and diseased living systems.