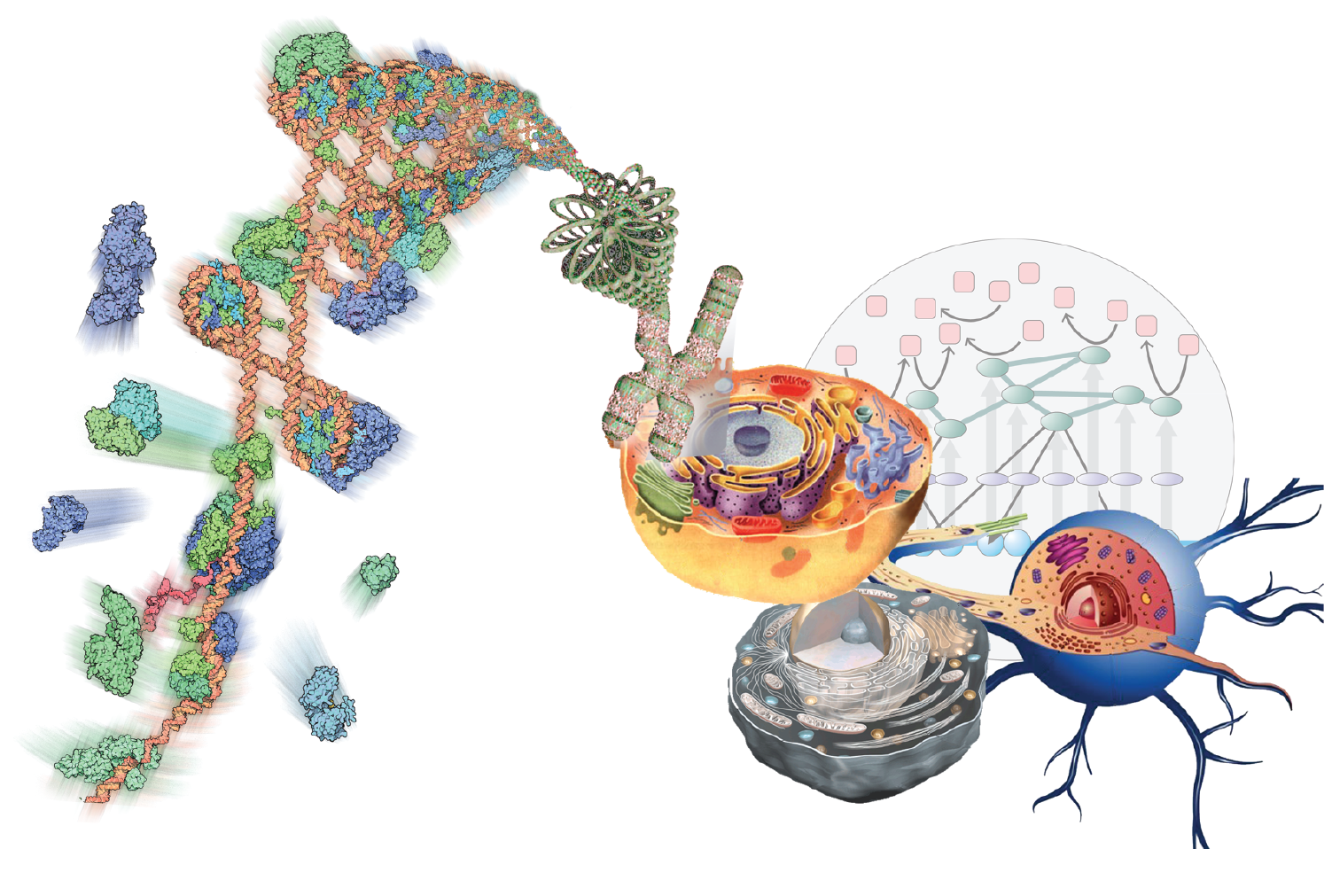
The Multiscale Molecular Microscope
TR&D Project 4. The Modeling Stage: Multiscale Spatiotemporal Modeling of Macromolecular Systems in Cellular Neighborhoods
Armed with the data generated through TR&Ds 1-3, we will interpret them through modeling. implementing them in our open-source Integrative Modeling Platform (IMP) package.
Our goal is to develop and support computational methods for building data-derived models of static and dynamic macromolecular structures as well as molecular networks representing cellular processes. We emphasize data on molecular interactions that favor quality over quantity and mechanism over scale. In the structural domain, the data include composition, stoichiometry, and connectivity by MS, distances from chemical cross-linking and various optical spectroscopies, images and density maps from EM, protein and domain contacts from affinity capture, shape from SAXS, as well as atomic structures from X-ray crystallography and NMR spectroscopy. In the network domain, the primary data-generating methods include qRT-PCR, microarray, RNA-Seq, ChIP-chip, ChIP-Seq, quantitative MS, fluorescence microscopy, pulse chase, flow cytometry, enzyme and ligand-binding assays.
We construct and advance each model in parallel with experimental biology. This close juxtaposition between modeling and experiment generates a cycle, where experiments set the initial parameters for a model that is then refined based on further experiments inspired by the model. The earlier a model can be generated, the more effective are the experiments and thus the overall process.
Developing tools for integrative structural biology.
Our integrative structure modeling approach casts the building of models as a computational optimization problem where knowledge about the assembly (TR&Ds 1-3) is encoded into the scoring function used to evaluate candidate models. The approach is implemented in Integrative Modeling Platform (IMP). In particular, we are working to improve the accuracy of ranking alternative models of static macromolecular structures; improve the search for assembly configurations that satisfy the data; apply the integrative approach to structural mapping of dynamic processes; and engage with others to develop an integrative structural biology toolbox of compatible representations and modules.
To learn more about IMP, click here…
As in structural modeling, we focus on details and mechanism.
Kinetic models have tremendous utility for understanding and predicting mechanisms of control and guiding experimentation; their simplicity enables ready adoption by the biomedical community; and can yield fundamental insights into numerous biological systems. A common perception in kinetic modeling is that one must have a wealth of highly time-resolved and detailed biochemical parameters before kinetic models can be used. This is often not the case; many system behaviors are determined primarily by network structure and the mechanisms in each process within the network rather than the fine-tuning of individual parameters. Thus, fundamental insights are possible with limited, but targeted, experiments. We are working to develop an integrative computational platform for quantitative kinetic modeling of dynamical biomolecular systems that will facilitate the exploration of dynamical system characteristics.