NCDIR researchers helped determine the molecular architecture of RNA polymerase II bound to the protein Gdown1, an essential transcriptional repressor that is specific to multicellular animals (metazoans). The labs at NCDIR performed the cross-linking with mass spectrometry readout (Chait Lab) and integrative structural modeling (Sali Lab) to complement the the cryo-electron microscopy map and biochemical data of Prof Robert G Roeder’s Lab, to elucidate the structural mechanism that Gdown employs to repress RNA pol II, an enzyme Prof Roeder discovered almost 50 years ago in 1969!
To read more, the full article can be found at: https://www.nature.com/articles/s41594-018-0118-5
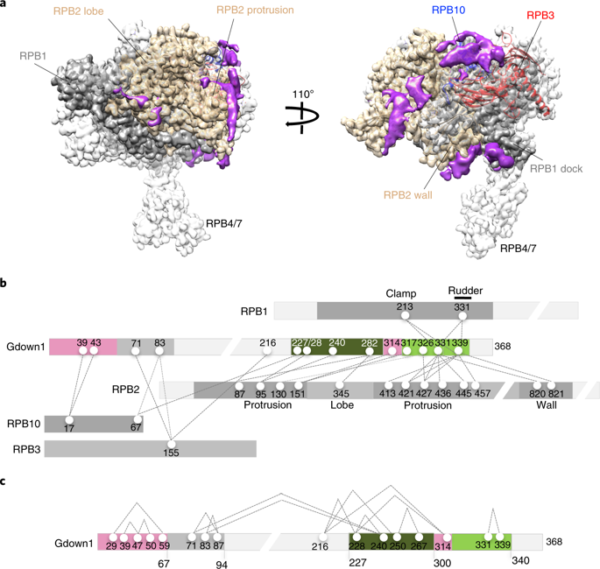
Fig. 1 | Molecular architecture of Pol II(G) by cryo-eM and CX-MS analyses. a, Cryo-EM structure of Pol II(G). Non-Pol II (Gdown1) density is shown in purple. An atomic model of human Pol II was obtained by real-space refinement of a model derived from relevant portions of the published model of bovine Pol II (PDB 5FLM). To identify Gdown1 density in the Pol II(G) map, the difference between all stable portions of the Pol II(G) cryo-EM map was calculated. b, Diagram of cross-links between Pol II subunits and Gdown1. Purified Pol II(G) was subjected to cross-linking with the amine-specific cross- linker, disuccinimidyl suberate (DSS), followed by high-resolution MS. c, Intra-molecular cross-links map of Gdown1. (from Jishage et al (2018). Nat Stuct Mol Biol)
