The nuclear pore complex (NPC) consists of nearly ~1000 protein of ~30 different types, and controls macromolecular transport between the nucleus and the cytoplasm. Extensive experimental efforts and integrative modeling have provided significant insight into the static structure of this complex, but molecular details underlying how this complex assembles has been lacking. An improved understanding of the two possible NPC assembly pathways may elucidate the crucial role the NPC has played in eukaryotic evolution and eventually decode the link between genetic alterations in NPC associated proteins and a variety of human diseases. However, the size of the NPC complex and timescales approaching one hour for assembly completion have posed challenges to previous studies.
In recent work, researchers from the labs of Jan Ellenberg and Andrej Sali worked together to investigate the molecular mechanism of NPC assembly. Using fluorescence correlation spectroscopy calibrated live imaging, protein composition is tracked throughout the assembly process. This stoichiometric analysis sufficiently distinguished the two known pathways for NPC assembly, and revealed that these pathways proceed through different mechanisms. Specifically, the order in which the central ring complex and nuclear filaments are inverted.
To gain more insight into the NPC assembly mechanism, the dynamic stoichiometry data and prior electron tomographs of assembly intermediates were integrated to create a molecular model for one of the assembly pathways, specifically the postmitotic assembly pathway. This spatiotemporal model predicts the structure of NPC intermediates throughout the assembly process, allowing for mechanistic predictions into postmitotic assembly. For example, our model suggests that the central channel complex likely helps dilate the membrane pore throughout the assembly process.
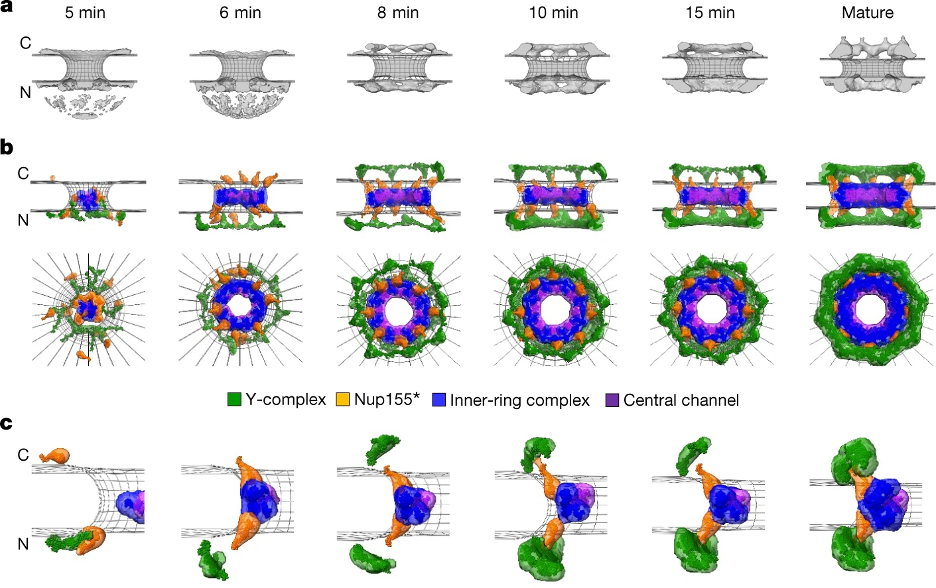
Integrative model of the postmitotic NPC assembly pathway. a, Protein density (grey) overlaid with the nuclear envelope surface (wireframe model, grey) at each time point used for integrative modelling. C, cytoplasm; N, nucleoplasm. b, The best-scoring model of the postmitotic assembly pathway (top and side views). The Y-complex is shown in green; an isolated fraction of Nup155 that is not forming a complex with Nup93, Nup188 and Nup205 is shown in orange; Nup93–Nup188–Nup155 and Nup205–Nup93–Nup155 complexes are in blue; and the Nup62–Nup58–Nup54 complex is in purple. c, The enlargement of one of the spokes.
Access the full text here!
