The Multiscale Molecular Microscope
TR&D Project 2. The Analysis Stage I: Tools for Analyzing the Composition and Stoichiometry of Macromolecular Assemblies
We will develop methods to faithfully and fully identify the bone fide components of any target macromolecular complex in the cell, as well as that of vicinal associators, with high accuracy and throughput.
Our overall goals are to be able to create and optimize methods to:
Goal 1: characterize any assembly in the cell – identifying its individual components
Goal 2: determine how many of those individual components make up the complex – the stoichiometry of the complex
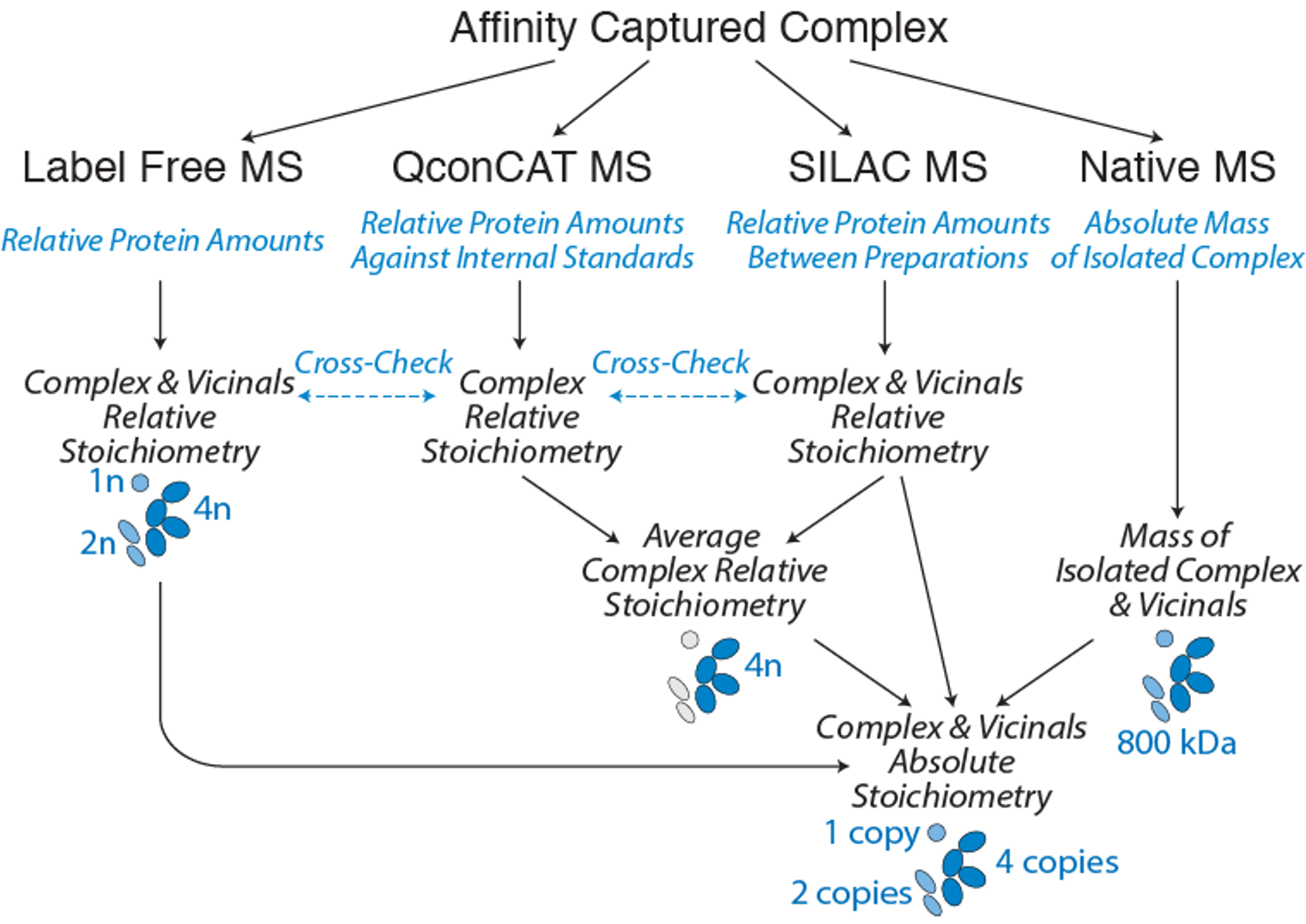
We aim to provide this technology to the community in the form of a “Stoichiometry Package”. This package will incorporate a parallel suite of techniques that give complementary and cross-verifying information, collectively providing accurate, comprehensive sets of stoichiometry data (and dynamic changes) for an isolated assembly. Users can choose any or a combination of approaches, depending on need.
Determining the Composition of Macromolecular Complexes and their Neighborhoods
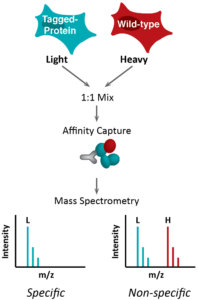
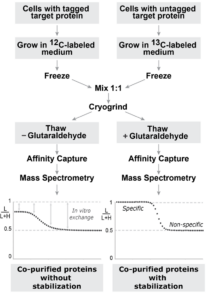
With high quality samples from Project 1, wear to characterize those complexes using variations of Isotopic Differentiation of Interactions as Random or Targeted (I‑DIRT), that involve enhancement using chemical cross linking (enhanced I-DIRT) and using a label-free method (No-DIRT). Additionally we will also analyze for any nucleic acid component of complexes.
I-DIRT pipeline (left) and enhanced I-DIRT (right)
To accurately determine the quantities of each interactor identified in the complex and how they change with time or assembly state we will use an optimized QConCat and various mass spectrometry (MS) based techniques (See image below). These techniques form the stoichiometry package we aim to develop for easy and accessible use to anyone interested in in-depth analysis of their complex of interest.