In collaboration with Charles Rice’s lab at Rockefeller University, NCDIR researchers have contributed to a new study that illuminates how a class of viruses called enteroviruses disrupt the machinery of normal human host cells during infection.
Enteroviruses cause a number of human diseases including gastroenteritis, the common cold, hand-foot-and-mouth disease, acute hemorrhagic conjunctivitis, and skin rash. In this study, researchers identified which host proteins are broken apart, or “cleaved”, by proteins produced by enteroviruses. By disrupting proteins this way, these viruses remodel infected cells to avoid detection by the host’s immune system and create a favorable environment for replication. The researchers identified proteins commonly targeted across five different enteroviruses and were able to identify specific cell functions in which these proteins participate, providing valuable insight into which cellular processes are subverted or disrupted during infection. As the first global assessment of enterovirus protein cleavage in human cells, the study is the first of its kind and provides a valuable community resource for understanding enterovirus biology and for generating new approaches to combat enterovirus-related diseases.
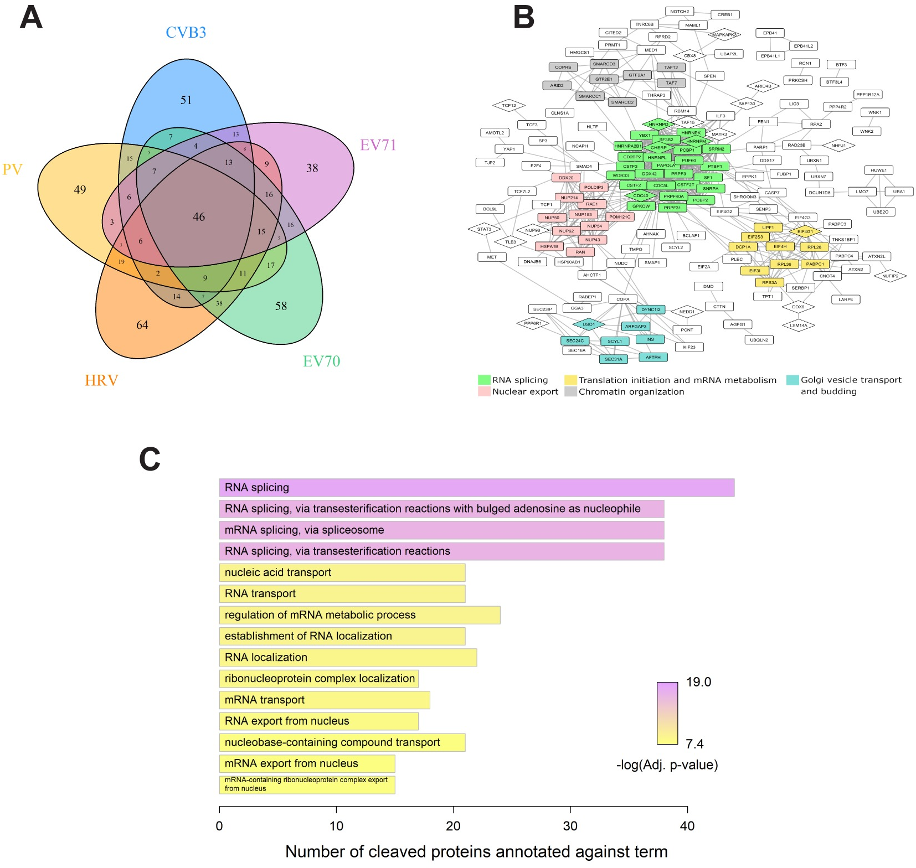
Figure: Analysis of proteins broadly targeted by enteroviruses.
(A) Venn diagram showing the numbers of cleaved proteins common or unique to different viruses. (B) STRING interaction network of proteins targeted by at least two of the five enteroviruses as rendered in Cytoscape (disconnected nodes are not shown). Coloring indicates groups of five or more proteins identified as highly interconnected by the MCODE clustering algorithm. Most significantly enriched biological processes are summarized below the network. Cleaved proteins validated by western blot are shown in diamond shaped nodes. (C) Top 15 significantly enriched biological processes targeted by at least two of the five viruses.
Full citation and article access:
Saeed M, Kapell S, Hertz NT, Wu X, Bell K, et al. (2020) Defining the proteolytic landscape during enterovirus infection. PLOS Pathogens 16(9): e1008927. https://doi.org/10.1371/journal.ppat.1008927