A native mass spectrometry (nMS)-based platform developed at the NCDIR for evaluating and optimizing sample quality for single particle cryogenic microscopy (cryo-EM) is featured in the current issue of the journal Structure (https://www.cell.com/structure/home).
The exponential increase in high-resolution biomolecular structures determined by cryo-EM is yielding unprecedented structural insights into cellular processes. Despite its extraordinary capabilities, one major bottleneck in single-particle cryo-EM involves sample preparation and the need to have an efficient means for assessing the quality of samples prior to committing the substantial resources, time and effort needed for a typical cryo-EM analysis. To address this need, Dom Olinares, a Research Associate in Brian Chait’s lab at the NCDIR worked with structural biologists to develop an nMS platform that readily integrates into the cryo-EM sample preparation workflow providing rapid feedback on sample integrity/purity and optimal preparation conditions.
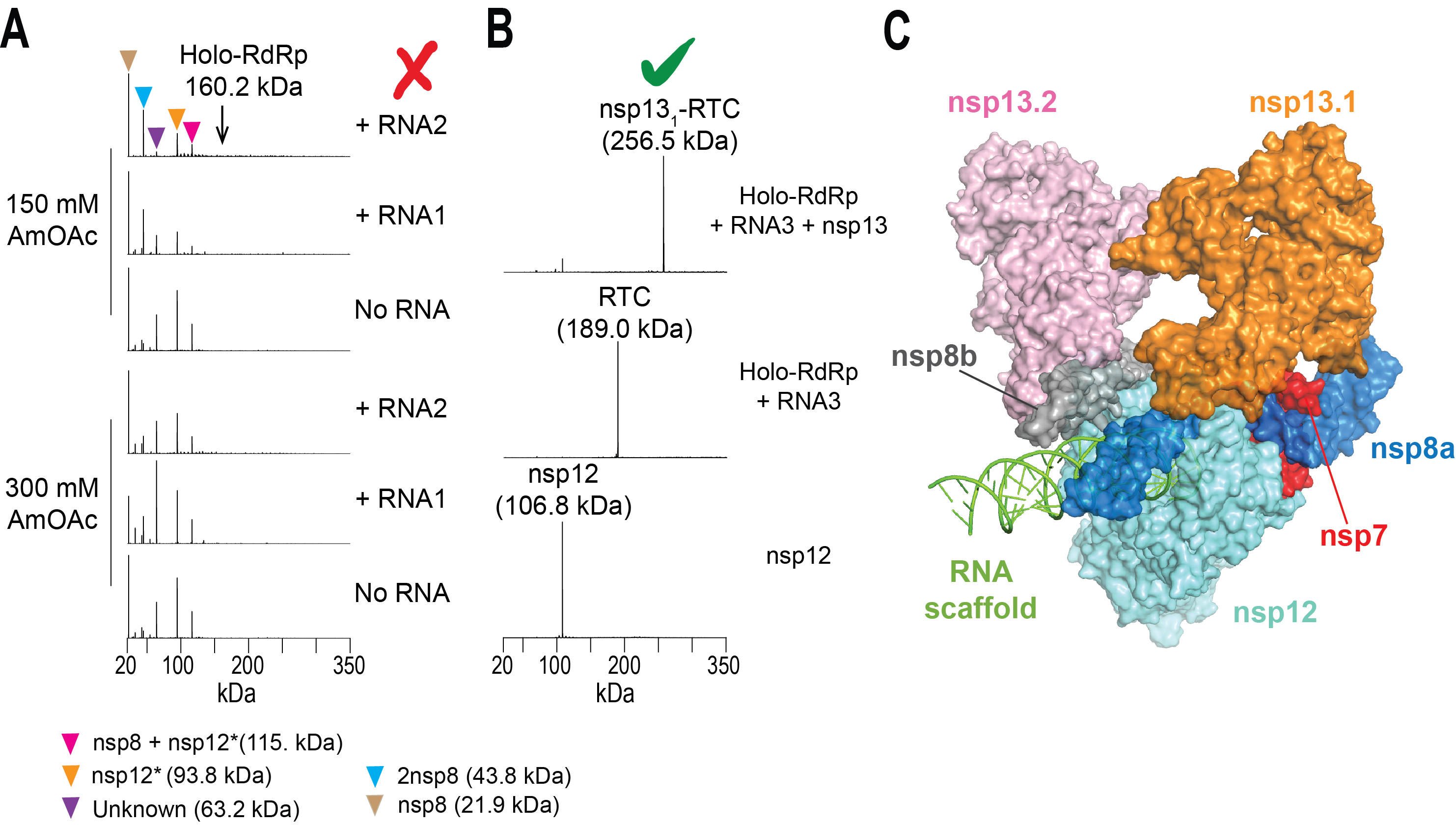
Figure. nMS-based screening to assemble the intact SARS-CoV-2 helicase-RTC. (A) Screening initial conditions for RTC with varying RNA scaffolds and ammonium acetate (AmOAc) concentrations did not form any assembled complex. Detection of unassembled subunits and a truncated nsp12 indicated issues with nsp12 expression. (B) nMS results after optimizing expression of the full-length nsp12 and subsequent reconstitution of RTC. Incubation of the assembled RTC with the nsp13 helicase yielded a single peak corresponding to nsp13-RTC at 1:1 stoichiometry. (C) cryo-EM structure of the nsp13-bound RTC (PDB ID: 6xez) (Olinares et. al., 2020).
Because nMS enables accurate mass analysis of intact protein complexes, it is well-suited for routine evaluation of the composition, integrity, and homogeneity of samples prior to their plunge-freezing on EM grids. In addition, once the EM data is collected, the nMS spectra acquired from the screening provide critical information on subunit composition, stoichiometry, protein modifications, and bound cofactors/ligands that serve as key inputs for EM structure reconstruction and validation.
The utility of the nMS-based diagnostic and screening platform was demonstrated through examples that characterized several bacterial transcription complexes as well as the replication-transcription complex (RTC) coupled to nsp13 helicase from SARS-CoV-2. Most of these structural projects started out with reconstitution conditions that did not yield the desired target assemblies for cryo-EM. In each case, with the aid of iterative nMS-based screening, it proved possible to rapidly establish optimized sample conditions that yielded cryo-EM structures at high resolution.
Publication:
Olinares PDB, Kang JY, Llewellyn E, Chiu C, Chen J, Malone B, Saecker RM, Campbell EA, Darst SA, Chait BT. “Native Mass Spectrometry-Based Screening for Optimal Sample Preparation in Single-Particle Cryo-EM.” Structure. 2020 Nov 17:S0969-2126(20)30413-5. PMID: 33217329.
The full text of the paper is freely available here:
https://www.cell.com/structure/fulltext/S0969-2126(20)30413-5
