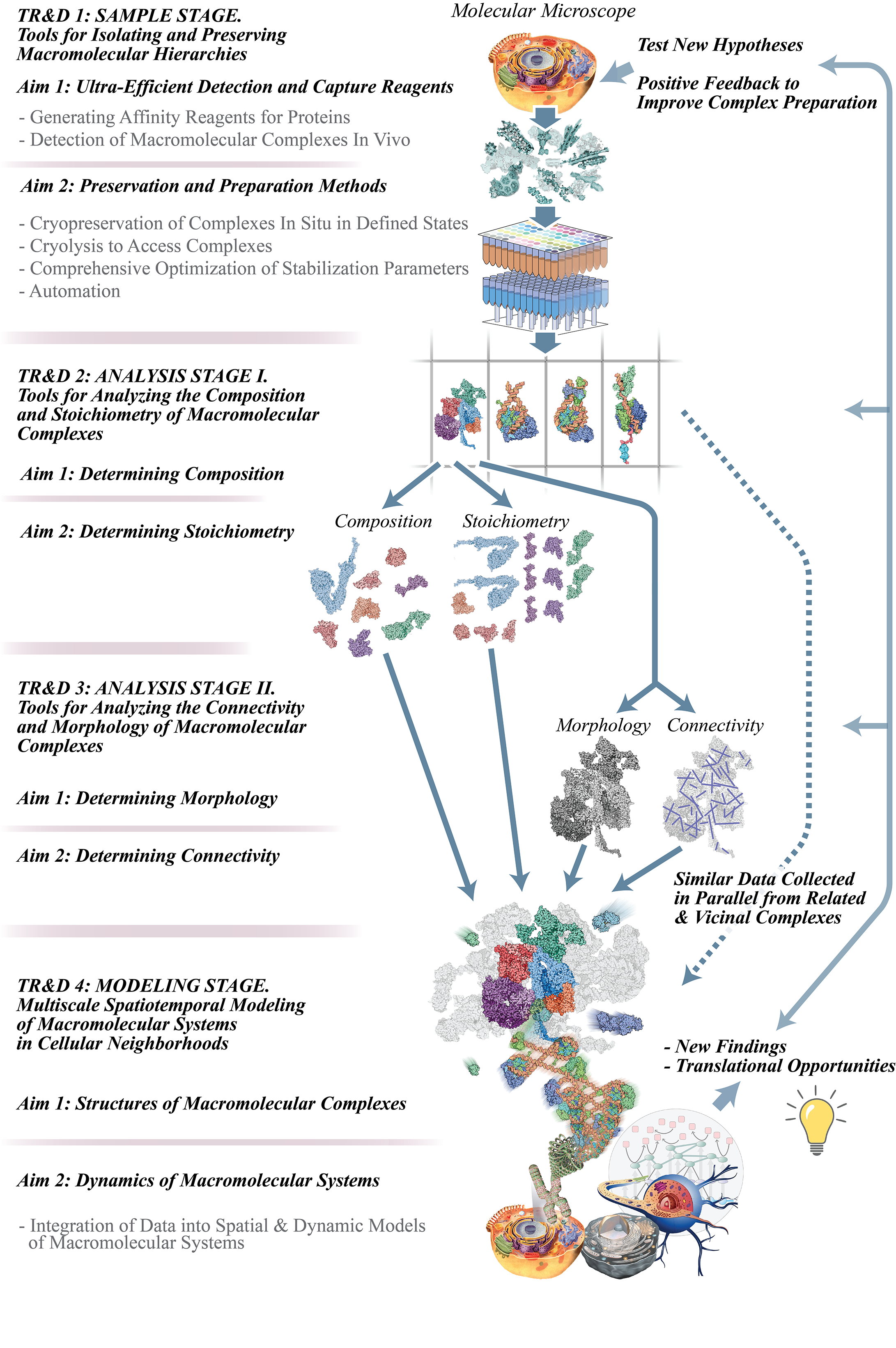
The Multiscale Molecular Microscope: an Interactomics Discovery Pipeline.
We ultimately seek to understand each interaction at atomic and biological time scales. This goal justifies our pipeline approach, consisting of TR&Ds 1 – 4 (see image and descriptions below) to respectively preserve, read, and interpret macromolecular interactions. One might think of our approach as a “multiscale molecular microscope”. In this analogy, TR&D1 prepares the samples for observation; TR&D2 and TR&D3 enable detailed observation and analysis of the sample; and TR&D4 enables integration of the data into a dynamic and interpretable picture of the sample. Our pipeline produces “classical anatomy at the molecular scale”, isolating and characterizing each component of the machine and ultimately contextualizing it in the specific living environment, so deriving that component’s specific structural functionality. The pipeline builds upon our existing NCDIR work, while incorporating a variety of new methods. It is modular, flexible, and adaptable, so that any investigator can enter and exit at any point, as required by their research priorities; and because of its modularity, researchers will be able to adopt those portions of the pipeline most pressing to their needs and continue to adopt other modules as their research advances.
TR&D Project 1 will develop technologies that purify and preserve, with high fidelity, various defined forms of the hierarchical arrangement of interactors surrounding any chosen macromolecule. We will address issues of signal-to-noise while faithfully preserving and isolating complexes in their native states, including components present in low abundance as well as those interacting dynamically.
TR&D2 focuses on advancing quantitative proteomics strategies to objectively define the makeup of isolated complexes, including innovative methods to trap transient and weak interactors, prevent post-lysis exchange, and reliably and easily distinguish specific from non-specific interactions. We are also assembling a practical “stoichiometry package” that integrates several cross-checked strategies to determine complex stoichiometry with high accuracy and determine state-specific changes.
TR&D3 will implement a fundamentally new concept of simultaneously generating complexes for cryo-EM and crosslinking-MS analyses that will revolutionize our ability to resolve large macromolecular structures to subnanometer levels, including membranous complexes and organelles. These methods will also enable the development of state-dependent stabilization and analysis methods to inform structural dynamics.
TR&D4 seeks to develop formal integrative methods for deriving structural and network models based on diverse experimental datasets generated by technologies in TR&Ds 1-3 – rather than models relying on single data sources, as is common now. In particular, the proposed developments will improve our ability to determine near-atomic resolution static structures for both homogenous and heterogenous samples; they will also allow us to compute dynamic models of multi-protein systems by satisfying a variety of spatiotemporal data.