Latest News from the NCDIR
The NCDIR’s Prof. Michael Rout presents at the SINGLE-DOMAIN ANTIBODIES 2023 Conference
September 18, 2023
The Single-Domain Antibodies 2023 meeting was held in Paris, September 18th-20th at the Conference centre (CIS) of the Institut Pasteur. It was the 3rd congress on single-domain Antibodies after Nanobodies 2019 in Bonn and Nanobodies 2021 in Brussels. This congress allows the scientific community to meet and share the latest advances in this fast-growing scientific field.
During this meeting, NCDIR director, Professor Michael Rout was invited to give a talk on “A Pipeline for Production of Nanobody Repertoires with Diagnostic and Therapeutic Potential” at the “Single domain Antibodies in Clinics: cancer and inflammatory diseases” workshop. Here is a brief summary of his talk.
Nanobodies are small single domain antibodies derived from camelids that can be targeted with high specificity against almost any antigen. Moreover, they can be inexpensively expressed in virtually unlimited quantities in simple bacterial cultures, as well as engineered to maximize their potential – they show tremendous promise as therapeutic reagents, because of their high specificity, small size and robustness.
We developed a mass spectrometry-based technology that overcomes the limitations of some other screening methods, enabling the rapid production of large repertoires of readily expressible recombinant nanobodies against any given antigen with very high affinities and specificities. We first applied this pipeline to the production of nanobodies that recognize common protein tags such as GFP, mCherry and TdTomato, to allow ultra-rapid capture of complexes for interactomics. In studying the structures of these nanobodies we have uncovered several alternative modes of epitope recognition virtually unique to this antibody class. During the COVID-19 pandemic, having recently refined and optimized our pipeline and supplemented it with a modified yeast display screening approach, we identified classes of nanobodies that have exceptional biophysical characteristics and unique and orthogonal mechanisms for neutralizing SARS-CoV-2, and that maintain their neutralizing efficacy against current variants; we are expanding this repertoire to generate nanobody formulations that will be able to neutralize any future betacoronaviral threats. We have also discovered strong synergistic effects between nanobodies, greatly enhancing their potency when used in combination. We are now using our pipeline to generate repertoires of nanobodies against other targets of biomedical importance, enabling use in CAR-T therapeutics, treatment or diagnosis of several cancer types, and various infectious diseases.
Publications related to Prof. Rout’s talk:
Ketaren NE, Mast FD, Fridy PC, Olivier JP, Sanyal T, Sali A, Chait BT, Rout MP, Aitchison JD. “Nanobody repertoire generated against the spike protein of ancestral SARS-CoV-2 remains efficacious against the rapidly evolving virus.” bioRxiv. 2023 Jul 14:2023.07.14.549041. doi: 10.1101/2023.07.14.549041. Preprint. PMC10369967
Cross FR, Fridy P, Ketaren N, Mast F, Li S, Olivier P, Pecani K, Chait BT, Aitchison JD, Rout MP. “Expanding and Improving Nanobody Repertoires Using a Yeast Display Method: Targeting SARS-CoV-2”. J Biol Chem. 2023 Jan 28;102954.doi: 10.1016/j.jbc.2023.102954. Epub 2023 Jan 28. PMC9884143
Mast FD, Fridy PC, Ketaren NE, Wang J, Jacobs EY, Olivier JP, Sanyal T, Molloy KR, Schmidt F, Rutkowska M, Weisblum Y, Rich LM, Vanderwall ER, Dambrauskas N, Vigdorovich V, Keegan S, Jiler JB, Stein ME, Olinares PDB, Herlands L, Hatziioannou T, Sather DN, Debley JS, Fenyö D, Sali A, Bieniasz PD, Aitchison JD, Chait BT, Rout MP. “Highly synergistic combinations of nanobodies that target SARS-CoV-2 and are resistant to escape.” Elife. 2021 Dec 7;10:e73027. doi: 10.7554/eLife.73027. PMC8651292
Fridy PC, Li Y, Keegan S, Thompson MK, Nudelman I, Scheid JF, Oeffinger M, Nussenzweig MC, Fenyö D, Chait BT, and Rout MP. “A robust pipeline for rapid production of versatile nanobody repertoires.” Nature Methods. 2014 Dec;11(12):1253-60. doi: 10.1038/nmeth.3170. Epub 2014 Nov 2. PMC4272012
Research Spotlight – Ultrasensitive detection of circulating LINE-1 ORF1p as a specific multi-cancer biomarker
A multi-lab team has worked with the NCDIR to make breakthrough developments in the realm of cancer biomarkers. The team engineering nanobodies – small, robust variants of monoclonal antibodies – for use in ultrasensitive and cost-effective pan-cancer biomarker assays. The research spotlighted LINE-1 as a uniquely specific marker for cancer, which, unlike in normal tissues, is evident not only in tissue but also in blood, bolstering its potential as an early detection tool and a prognostic metric.
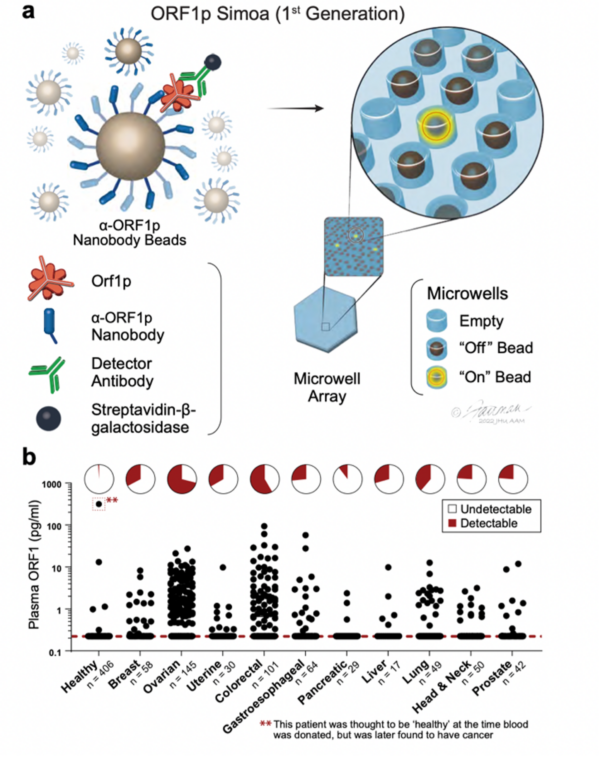
Highly Specific Detection of Carcinomas with the First-Generation ORF1p Simoa Assay. a, Schematic of single-molecule protein detection by Simoa; a first-generation assay is shown. Antibody/nanobody-coated magnetic beads, present in excess relative to target, capture single target ORF1p molecules; in the first-generation assay, beads are conjugated with α-ORF1p capture nanobody 5 (Nb5). Enzyme-labeled α-ORF1p detection reagent (here, an antibody, Ab6) is added, forming an “immunosandwich”; the beads are then loaded into microwells that each can hold at most one bead, and ORF1p molecules are then digitally detected using a fluorogenic substrate by counting “on” wells. b, First-generation ORF1p Simoa detects plasma ORF1p with high specificity across major carcinomas. Pie charts indicate percentage of samples with detectable levels; dashed red line, limit of detection.
Emphasizing collaboration, the study harnessed the dual expertise of NCDIR in producing specialized nanobodies and state-of-the-art mass spectrometry assays, and leverages a single-molecule-based detection technology known as SIMOA (developed by one of the team labs.
The test aims to identify the open reading frame 1 protein (ORF1p), a protein expressed by the mobile element LINE-1. High ORF1 expression is prevalent in numerous cancers, and signals a heightened risk of fatal cancer. Thus, for years, scientists have pursued a precise and sensitive method to detect ORF1p. The team characterized their research as an initial trial that exceeded their expectations, subsequently enhanced by a sequence of engineering refinements. The team utilized sophisticated immunohistochemistry stains, revealing a more widespread presence of LINE-1 in colon and gastroesophageal cancers. To counter the challenge posed by the extremely low LINE-1 levels in blood, they designed a cutting-edge SIMOA assay, using a combination of nanobodies and antibodies.
This assay was not only highly specific but also flagged an individual who was initially deemed healthy but later diagnosed with advanced prostate cancer. The team further refined their assays, producing 2nd and 3rd generation versions using custom nanobodies and antibodies, enhancing detection rates exponentially. These optimized assays demonstrated commendable accuracy in detecting early-stage lung and ovarian cancers and showcased their prognostic value, especially in the context of gastroesophageal and colon cancers. Notably, these assays can detect therapy response in gastroesophageal cancer earlier than other metrics. Plus, they stand out for their potential affordability, rapid results, and minimal blood requirement., making them excellent candidate assays for widespread clinical use.
Related publication:
Taylor MS, Connie W, Fridy PC, Zhang SJ, Senussi Y, Wolters JC, Cheng WC, Heaps J, Miller BD, Mori K, Cohen L, Jiang H, Molloy KR, Norden BL, Chait BT, Goggins M, Bhan I, Franses JW, Yang X, Taplin ME, Wang X, Christiani DC, Johnson BE, Meyerson M, Uppaluri R, Egloff AM, Denault EN, Spring LM, Wang TL, Shih IM, Jung E, Arora KS, Zukerberg LR, Yilmaz OH, Chi G, Matulonis UA, Song Y, Nieman L, Parikh AR, Strickland M, Corcoran RB, Mustelin T, Eng G, Yilmaz ÃH, Skates SJ, Rueda BR, Drapkin R, Klempner SJ, Deshpande V, Ting DT, Rout MP, LaCava J, Walt DR, Burns KH. “Ultrasensitive detection of circulating LINE-1 ORF1p as a specific multi-cancer biomarker.” Cancer Discov. 2023 Sep 12. doi: 10.1158/2159-8290.CD-23-0313. Online ahead of print. PMID: 37698949
The paper can be freely accessed here: https://aacrjournals.org/cancerdiscovery/article/doi/10.1158/2159-8290.CD-23-0313/729035/Ultrasensitive-detection-of-circulating-LINE-1
Research Spotlight – Nuclear pore complexes mediate subtelomeric gene silencing by regulating PCNA levels on chromatin
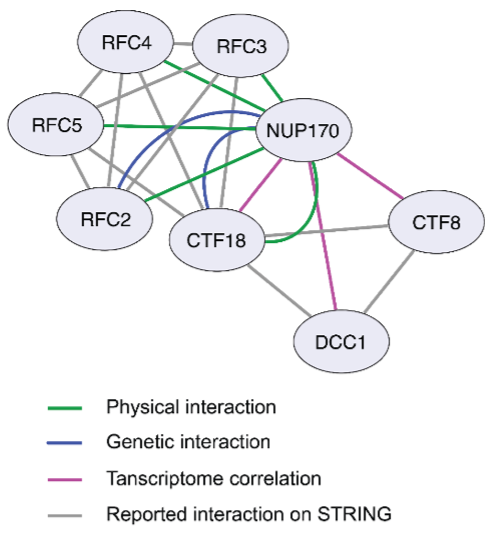
Figure 1: Cytoscape network illustrating the interaction between Nup170 and the Ctf18-RFC complex.
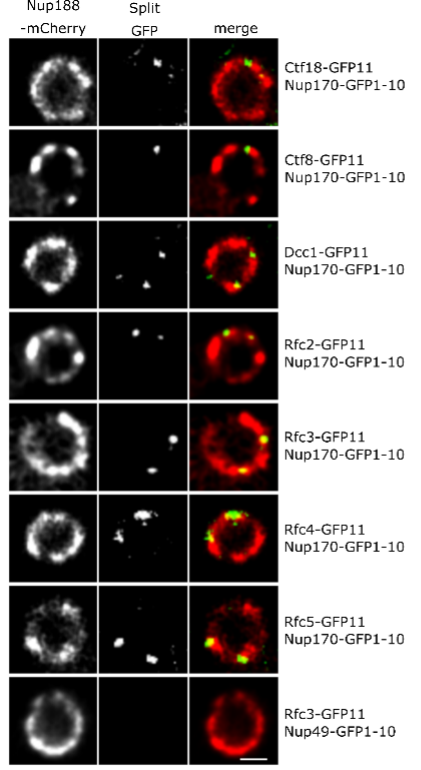
Figure 2: Ctf18-RFC is recruited to a subset of NPCs lacking the nuclear basket nucleoporins Mlp1 and Mlp2.
The Nuclear Pore Complex (NPC) functions as a crucial gateway, mediating the transport of macromolecules between the nucleus and the cytoplasm, thus playing a central role in regulating gene expression. In addition, the NPC’s physical interaction with chromatin significantly affects transcription, chromatin structure, and DNA repair. For example, the interaction of active genes with the NPC propels transcription, while interaction with telomeres fosters transcriptional silencing. The intricacies of these interactions, requiring different core channel proteins like Nup170, underscore the multi-faceted roles of NPCs in cellular functions. Mutations in NPC components can be deleterious, and potentially linked to a myriad of diseases, thereby amplifying the importance of in-depth structure-function studies of the NPC.
Our recent work, published in the Journal of Cell Biology (JCB), utilized an integrative systems cell biology approach encompassing genomic, proteomic, transcriptomic, and biochemical methods to provide mechanistic insight into how Nup170 facilitates its regulatory role over gene silencing. As part of our analysis, we compared the transcriptome signature of cells lacking Nup170 to a compendium known as ‘the deleteome’, which contains the transcriptomic profiles of approximately 1,500 individual yeast gene deletion strains. We pinpointed the deletion strain of Replication Factor C (RFC) complex member Chromosome Transmission Fidelity 18 (ctf18Δ) as having a similar transcriptomic signature as the nup170Δ strain (Figure1). Additionally, RFC complex members physically associate with Nup170, as identified by affinity purification mass spectrometry methods.
What is the significance of these observations? To our surprise, we found that Nup170 has a role in the Replication Factor C (RFC) complex-mediated loading of Proliferating Cell Nuclear Antigen (PCNA) onto DNA. PCNA serves as a sliding clamp to enhance the processivity of DNA polymerase during DNA replication and repair, but it also acts as a versatile hub in a myriad of additional cellular processes, including DNA mismatch repair, chromatin remodeling, and cell cycle regulation. Notably, upon Nup170 loss, a genome-wide decrease in PCNA levels was observed, as opposed to a telomere-confined decrease, suggesting a broader, systemic role of the NPC in mediating PCNA loading functionality. Furthermore, our experiments revealed that this functionality of Nup170 is specific to a subset of NPCs that lack a nuclear basket (Figure 2). To determine if PCNA levels are a critical factor in controlling telomeric silencing, we combined the nup170Δ with a PCNA unloading factor mutant (elg1Δ). Indeed, combining the loss of Nup170 with the loss of Elg1 resulted in increased PCNA levels and we also observed normal transcriptional silencing of sub-telomeric genes. Thus, increased PCNA levels on chromatin by the inactivation of an unloading factor is sufficient in maintaining telomeric silencing. These findings represent a leap forward in understanding the functional specialization of NPCs, specifically, the role of Nup170 in DNA damage repair and heterochromatin formation (Figure 3).
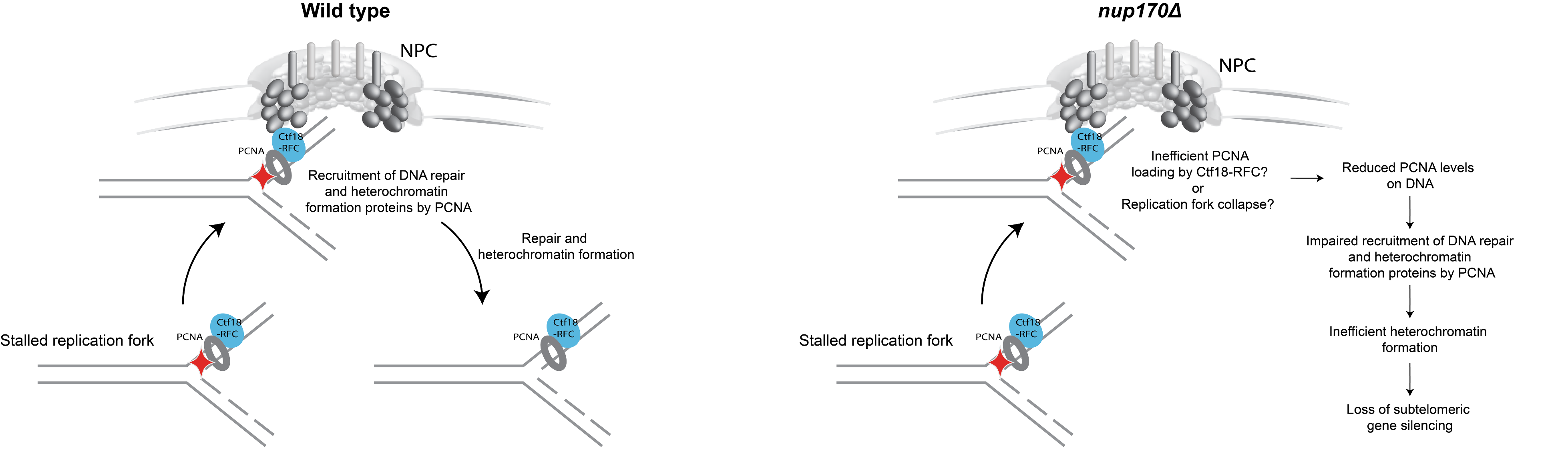
Figure 3: Model illustrating the role of the NPC in DNA damage repair and heterochromatin formation.
Publication:
Kumar S, Neal ML, Li S, Navare AT, Van Eeuwen T, Wozniak RW, Mast FD, Rout MP, Aitchison JD. “Nuclear pore complexes mediate subtelomeric gene silencing by regulating PCNA levels on chromatin.” J Cell Biol. 2023 Sep 4;222(9):e202207060. doi: 10.1083/jcb.202207060. Epub 2023 Jun 26. PMC10292210
The paper can be freely accessed here: