NCDIR scientists contributed to screening and providing key information in capturing the the SARS-CoV-2 replication transcription complex (RTC) bound to either its natural substrate or with the antiviral remdesivir yielding insights into how the drug works.
SARS-CoV-2 relies on the replication and transcription of its genetic material in order to spread and cause disease. This process involves a multi-protein assembly called the RTC, consisting of the RNA-dependent RNA polymerase and accessory proteins that are bound to RNA. The process begins with the binding of an incoming nucleotide triphosphate (NTP) into the active site of the RTC followed by the catalytic incorporation of the NTP into the elongating RNA product. As this essential process repeats many thousands of times per one round of replication/transcription, it is vulnerable to inhibition by substrate analogues which could introduce sequence errors or lead to premature termination. Indeed, two antivirals (remdesivir and molnupiravir) that target the RTC are currently being used in the clinic for treating COVID-19.
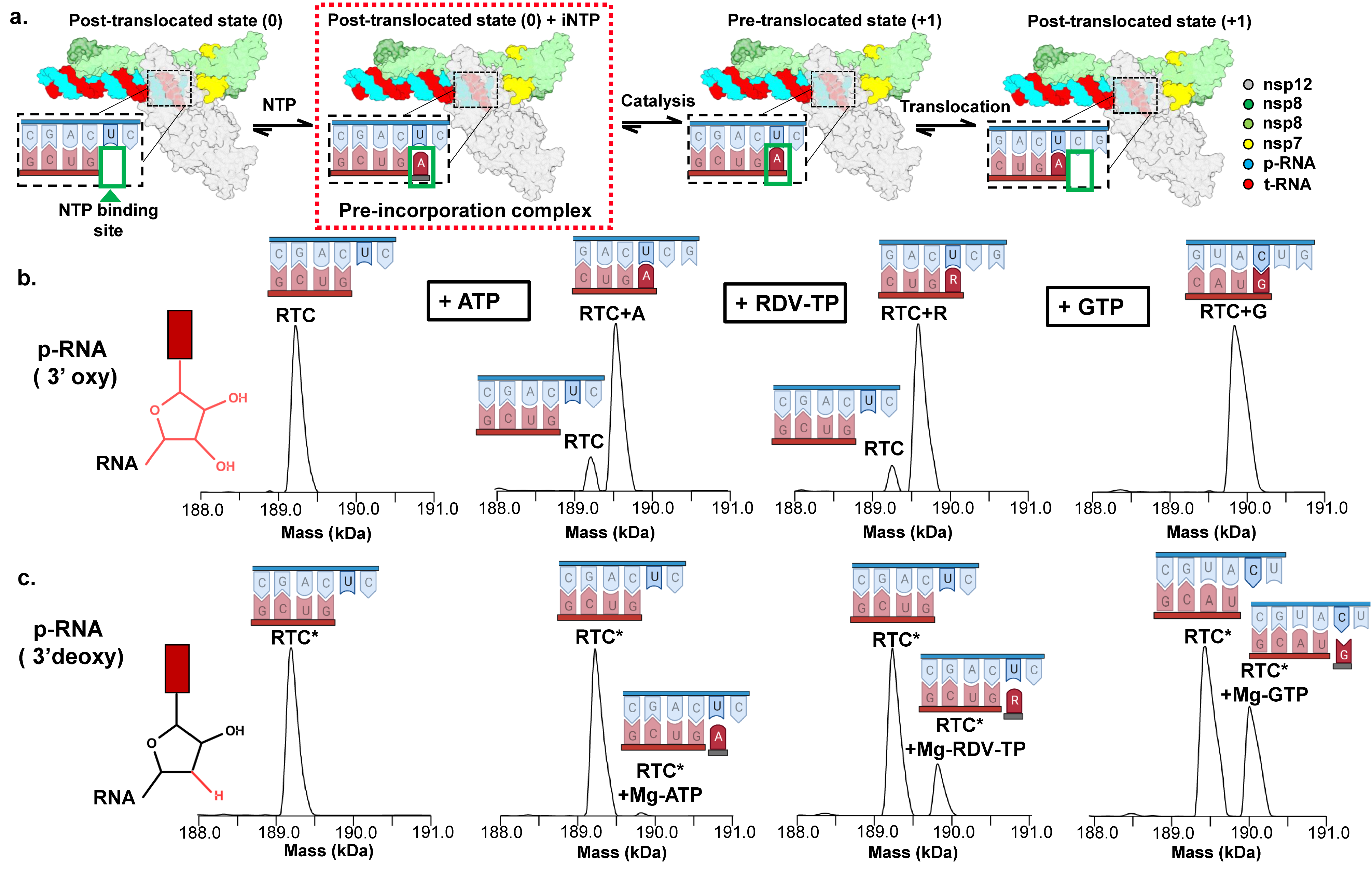
Figure 1: Native MS-based screening for RTC pre-incorporation complexes. (a) Schematic depicting the major steps of the nucleotide addition cycle of the RTC. The pre-incorporation complex studied here is highlighted (red dashed box). (b) nMS analysis of the RTC bound to a 3’oxy p-RNA in the absence and presence of NTPs. (c) Similar nMS analysis to (b) but the RTC was reconstituted using a 3’-deoxy p-RNA (RTC*) (Malone et. al., 2023).
To better understand how the RTC recognizes and differentiates among the various NTP substrates and the drug remdesivir, the Darst and Campbell lab at The Rockefeller University and a team of scientists from Gilead Sciences, the drug company that developed remdesivir, sought to capture the state where the incoming NTP substrate is bound by the RTC prior to catalysis, also known as the pre-incorporated state. Guided by previous work on other polymerases, the group investigated a range of chemical strategies to block incorporation after substrate binding using the native mass spectrometry (nMS)-based screening platform that was developed at the NCDIR. This platform has become an essential component of the lab’s structural biology workflow and has reliably provided a wealth of critical information in preparing samples for successful single-particle, cryo-EM analysis.
According to Dom Olinares, a research associate from Brian Chait’s lab who developed the platform:
“nMS proved such a valuable tool because this method enabled accurate and high-resolution mass measurements of intact RTCs and preserved the critical noncovalent interactions within the RTC. We could clearly distinguish from the mass profiles whether the added NTP was covalently incorporated to the growing RNA or was just sitting in the active site waiting to react.”
The most successful strategy for stalling the reaction was by using a product RNA strand with a 3’-deoxy end instead of the canonical 3’-oxy end. This blocked the reaction of the incoming NTP substrate but still allowed the substrate to remain in the active site (Figure 1). Overall, the group was able to successfully obtain cryo-EM structures that visualized the RTC bound to each of the natural NTPs in states poised for incorporation (Figure 2) as well as determined the structural basis for the selective incorporation of remdesivir over its natural counterpart ATP (Figure 3). The results explain the suite of interactions required for NTP recognition and inform the rational design of new antivirals that target the coronavirus RTC.
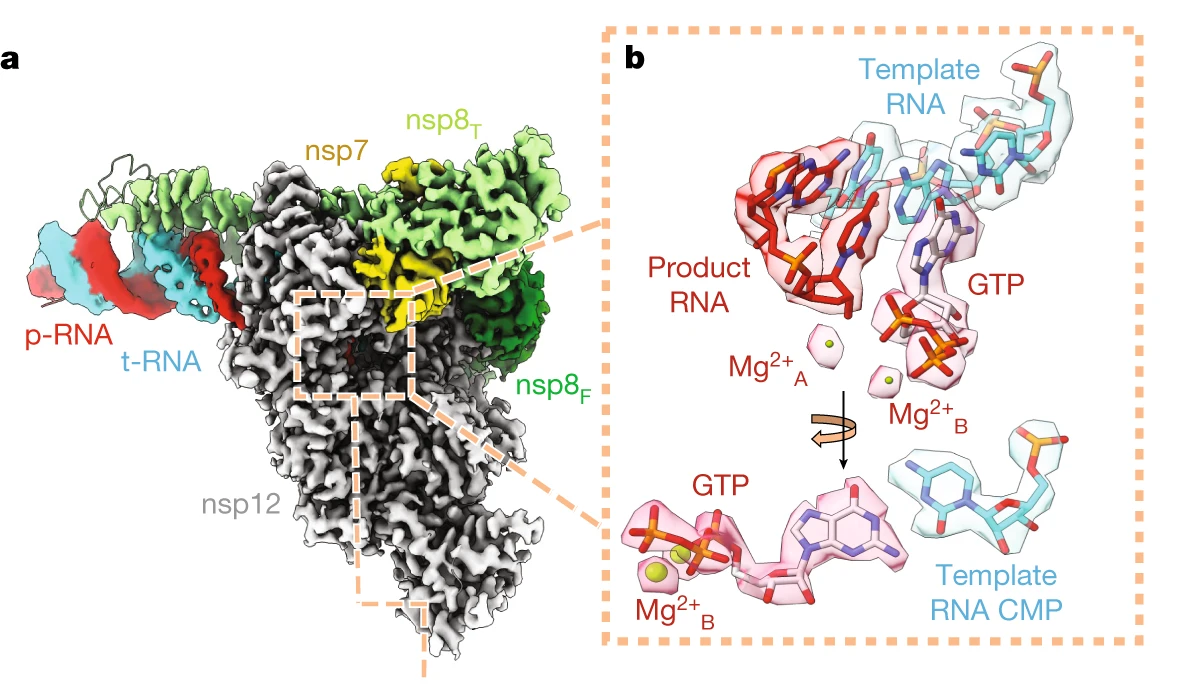
Figure 2: Representative cryo-EM structure of an RTC pre-incorporation complex. (a) Cryo-EM density for RTC+GTP structure. (b) Zoom-in of the active site showing the bound incoming GTP, two associated metal ions and the nearby bases of the t-RNA and p-RNA strands (Malone et. al., 2023).
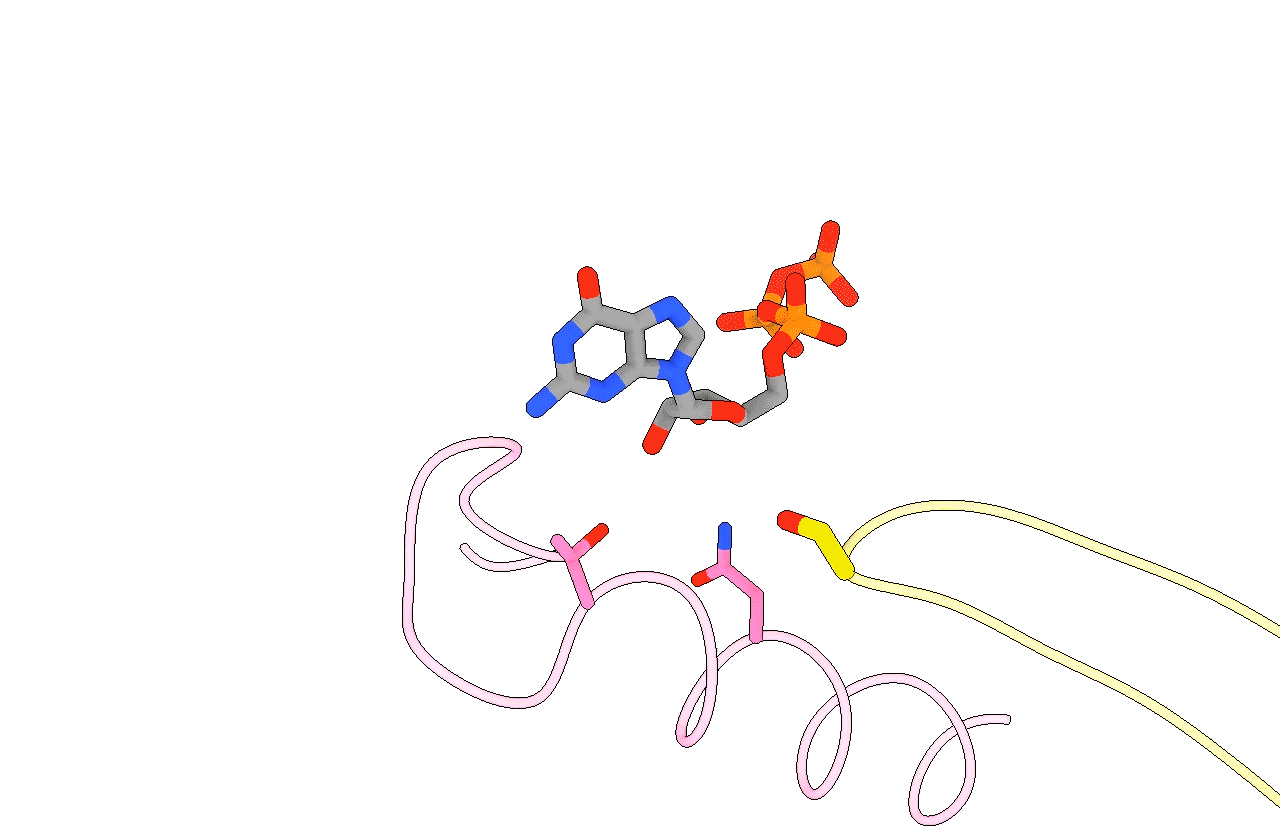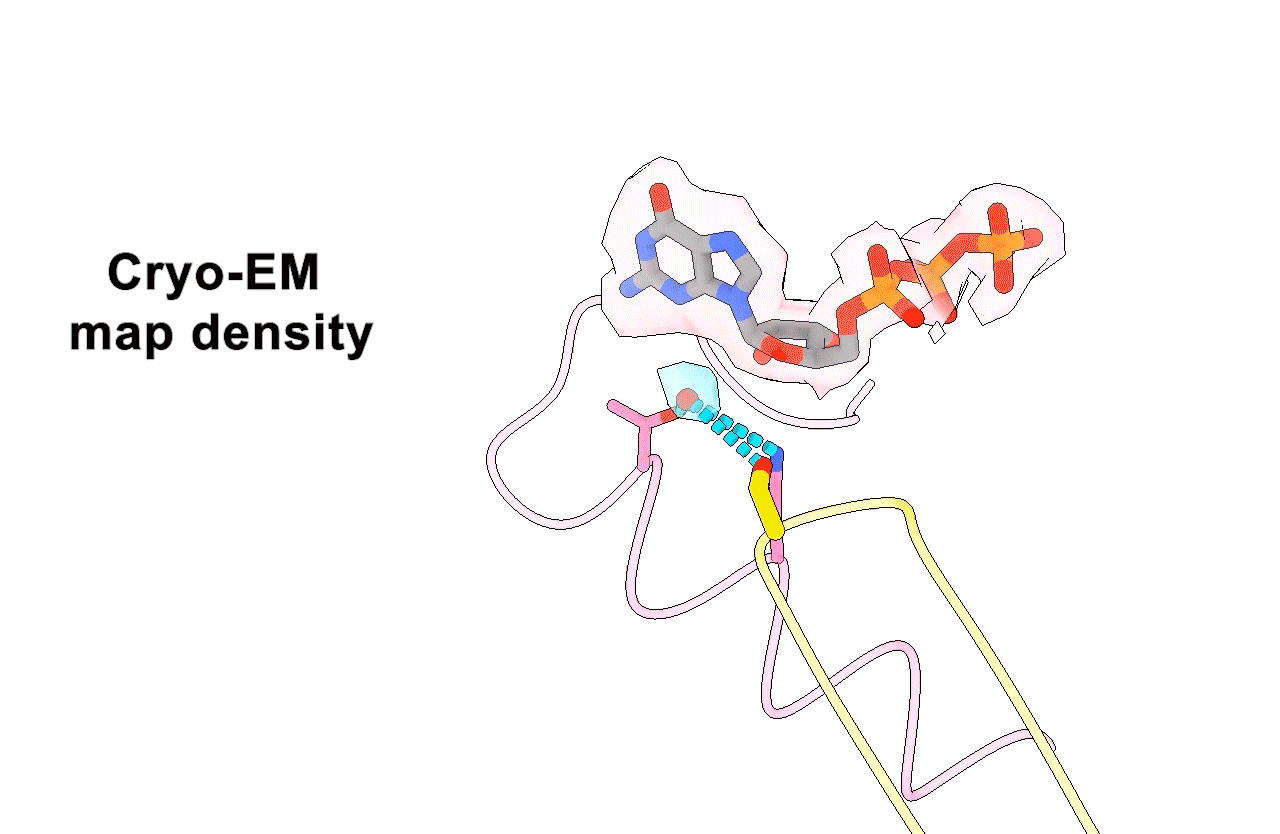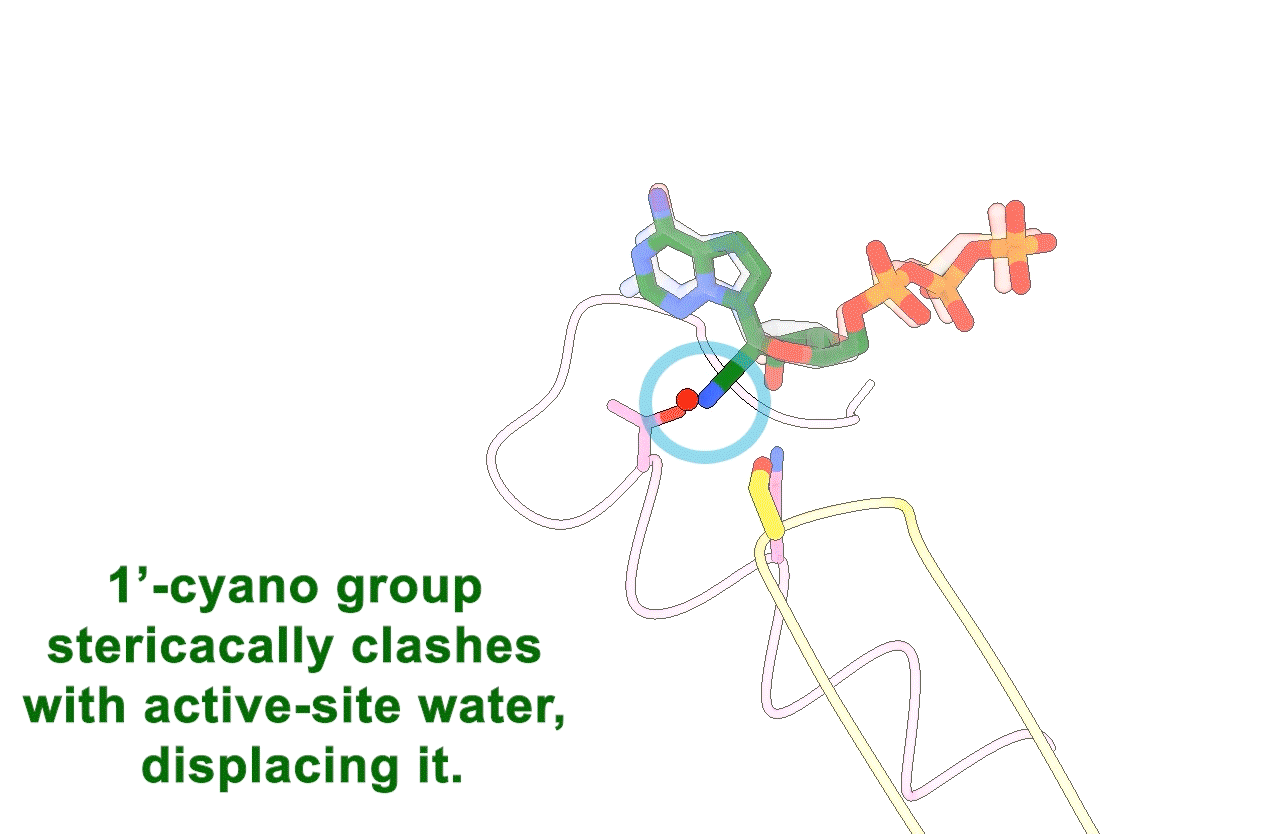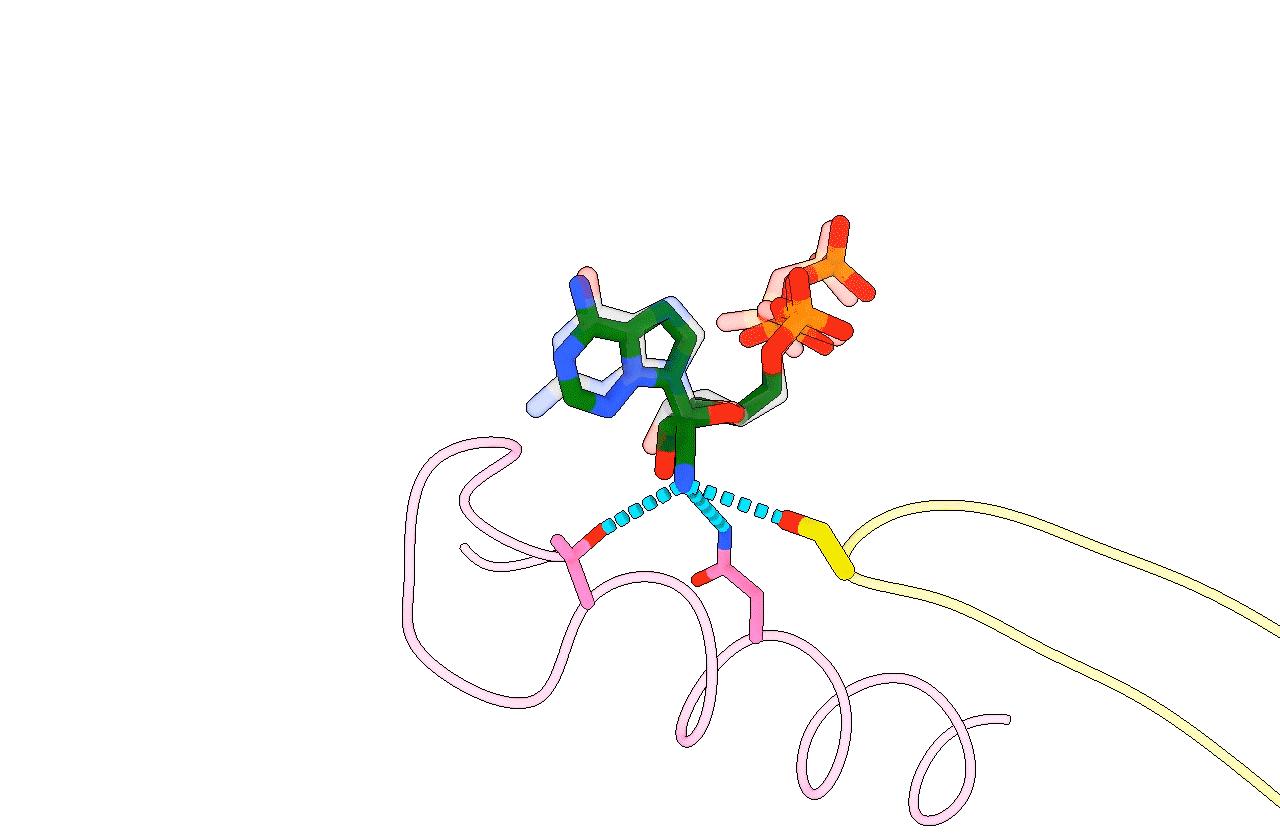
Figure 3. Animation highlighting the key differences in active site interactions between the RTC’s natural substrate compared to remdesivir.
Publication:
Malone BF, et. al. Structural basis for substrate selection by the SARS-CoV-2 replicase. Nature. 2023 Feb 1:1–7. doi: 10.1038/s41586-022-05664-3. PMC9891196.
The paper can be freely accessed here: https://www.nature.com/articles/s41586-022-05664-3
