A new publication by NCDIR scientist Dr. John LaCava (RU) in collaboration with Dr. Kathleen Burns (Johns Hopkins U) and Dr. David Fenyö (NYU) reveal the difficulty of identifying key players in human cancers.

Dr. John LaCava

Dr. Kathleen Burns

Dr. David Fenyö
Retrotransposons are ’selfish’ DNA sequences that have the ability to replicate themselves in host genomes via a ‘copy and paste’ mechanism. That is: the gene is transcribed and the RNA is later reverse transcribed (i.e. retro) into a new cDNA copy that is inserted pasted into a new location in the host genome (i.e. transposed). As a result of this continuous copy and paste proliferation, over evolutionary time LINE-1 retrotransposon DNA sequences have come to compose ~20% of the human genome (~1/2 million copies). Although the vast majority of these sequences are ancient and defunct, up to ~300 such loci may be capable of active proliferation, particularly during gametogenesis and oncogenesis. Each active LINE-1 can express two proteins: ORF1p and ORF2p that are both necessary but not sufficient for retrotransposition. ORF1p and ORF2p assemble together into ribonucleoprotein with the L1 RNA that encoded them (known as cis preference). Additional essential activities are provided by host proteins that L1 RNPs recruit and co-opt.
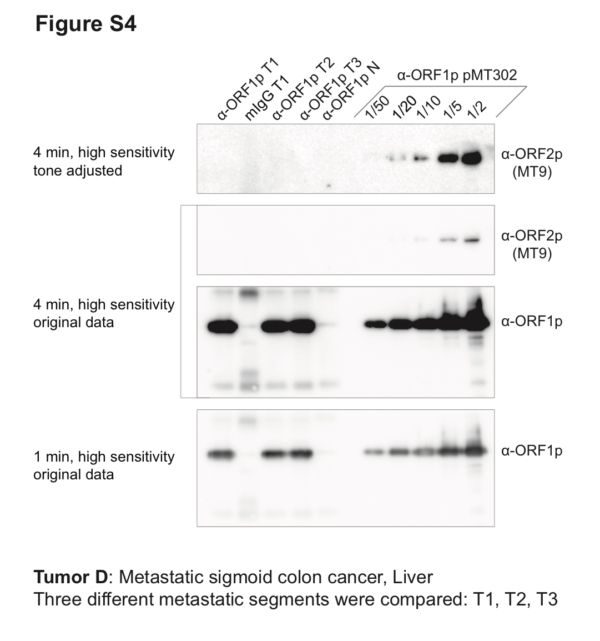
The proliferation of LINE-1 retrotransposons is apparent at the genetic level in cancers and cell lines – providing unequivocal genetic evidence for the expression of the enzymatic, replicative component of LINE-1: ORF2p. However, ORF2p continues to elude direct detection. In a just published study in Mobile DNA, three research teams (Fenyö, Burns, LaCava) – with further support from the NCDIR join forces and deep dive into data from the NCI’s Clinical Proteomic Tumor Analysis Consortium in search of ORF2p peptide signatures. Failing to find convincing evidence in the archived data, the researchers raised a collection of monoclonal antibodies against the protein in an effort to bolster capture and detection. Although the antibodies validated excellently against the target in ectopic expression, endogenous ORF2p remained elusive. Finally, the researchers took to affinity isolating endogenous LINE-1 RNPs, via ORF1p, directly from resected patient colorectal cancers – reporting the first endogenous LINE-1 cancer interactome. Although ORF2p was not identified among the co-isolated proteins, the researchers demonstrate the technical feasibility of studying disease-related protein complexes directly in patient tumors and identified overlaps between endogenous LINE-1 colorectal cancer protein interactions and those previously described from ectopically LINE-1 expressing HEK-293T cells. The researchers conclude that more sensitive methods will be needed to robustly detect ORF2p and propose targeted mass spectrometry and proximity ligation immuno-fluorescence as two future directions. The follow-up studies are currently under way.
Co-IP/Western blot. Three different segments of metastatic sigmoid colon cancer (from liver) were used as starting material for anti-ORF1p affinity isolations (α-ORF1p T1–3), including a mock-capture control using mouse IgG affinity medium with tumor extracts (mIgG T1), and matched normal tissue with anti-ORF1p affinity medium (α-ORF1p N). Co-IP of ORF1p/2p ectopically expressed from pMT302 in HEK-293TLD is provided as a comparative positive control. All co-IPs used 100 mg cells or tissues as input. 100% of the co-IP elutions done using patient tissues were analyzed; in contrast, fractions (labeled) of the co-IP from pMT302 in HEK-293TLD were analyzed. ORF1p yields from tumor were comparable to those obtained from 1/5th – 1/10th of a co-IP from pMT302/HEK-293TLD. However, while ORF2p signal is clearly detectable in 1/5th and closer to the baseline (but still eminently detectable) in 1/10th of a pMT302/HEK-293TLD co-IP, no ORF2p signal was observed in tumor co-IPs.